The Open Bioinformatics Foundation (OBF) Event Fellowship program aims to promote diverse participation at events promoting open-source bioinformatics software development and open science practices in the biological research community. David Kiragu Mwaura,_an Assistant Research Scientist at the Kenya Institute of Primate Research, was awarded an OBF Event Fellowship to attend the 2025 AIBBC conference.
Introduction
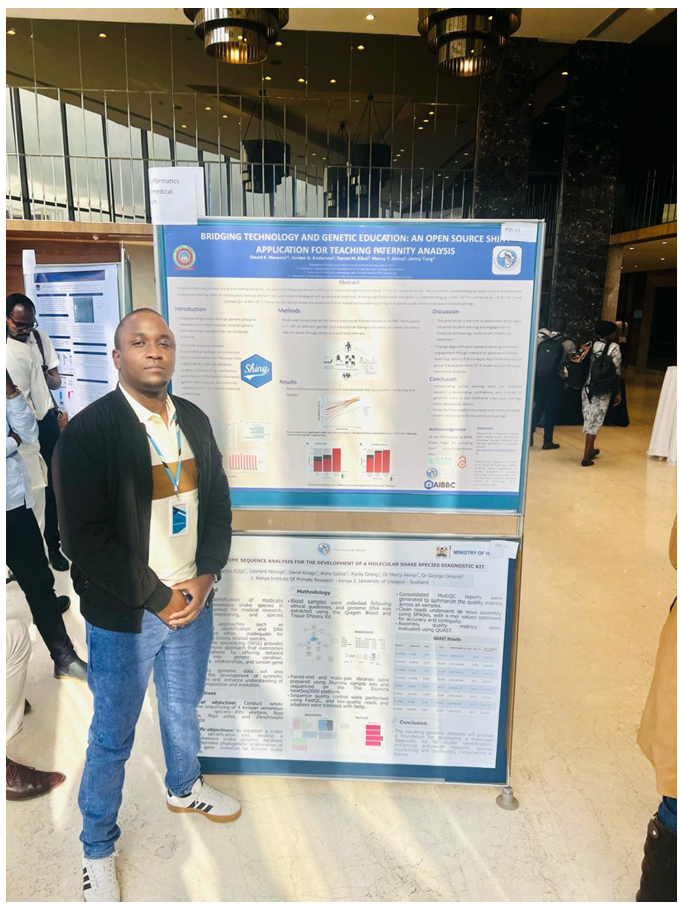
My desire to share my masters’ research output in a real world demonstration had finally come to fruition. This was achieved by securing a competitive travel grant from the Open Bioinformatics Foundation Event Fellowship. Leaving the chilly landscapes of Glasgow, Scotland for the warm, tropical weather in my home country, Kenya, I was honored to attend the 2025 African International Biotechnology and Biomedical Conference, where scientists from around the world gather to exchange ideas that directly address Africa’s needs.
[Read More]






 My team (Group 4) from the 2025 ESIIL Innovation Summit.
My team (Group 4) from the 2025 ESIIL Innovation Summit.